|
|
| Home | Current Issue | Archives | Bookshelf | Online Features | Marketplace | Subscribe |
| In This Section |
| Site Search |
| Visitor Login |
DOI: 10.1511/1998.3.216
COMPUTING SCIENCE
Prototeins
|
Anyone who has ever struggled to fold a roadmap should have an extra measure of respect for protein molecules, which fold up all on their own and practically put themselves away in the glove box. Protein folding is so remarkably efficient that it has been called a paradox. Thirty years ago Cyrus Levinthal pointed out that a typical protein molecule has so many possible configurations that it would need eons to explore all of them and find the best shape; yet proteins fold in seconds. Looking at the tangled loops and coils of a folded protein, you might imagine that the arrangement is haphazard—like a randomly crumpled map rather than a properly folded one—but in fact every twist and turn is precisely specified. Chemically, a protein is a linear polymer, a sequence of the smaller molecules called amino acids, which are joined end to end like pop-beads. The sequence of amino acids is the only information about the protein encoded in the genes, but the protein can do its job only if the one-dimensional chain of amino acids folds into the correct three-dimensional structure. Apparently the sequence alone is enough to guide the folding. If two protein molecules have the same sequence, they fold up into the same shape. One way to gain a better appreciation of the protein molecule's knack for folding is to simulate it with a computer program. The most detailed simulations track the motion of every atom and try to reproduce all the chemistry and physics going on in the system. The ultimate goal is to predict the native structure of the protein based on nothing more than the sequence of amino acids. Unfortunately, that goal is a distant one. The models require hours of computer time just to simulate a few picoseconds of molecular dynamics. I have been exploring a protein model at the other end of the complexity scale—a minimalist model, where every aspect of the simulation is reduced to its simplest possible form. A model so abstract cannot reveal anything about the structure of particular protein molecules—it cannot show how insulin or myoglobin folds—but it may offer clues to some general principles of protein folding. For example, one might hope to learn what kinds of amino acid sequences lead to a stable and compact molecule. The great advantage of a really simple model is that you can solve it exactly, at least for short chains of amino acids. You can examine every possible folding of every possible sequence, picking out the ones of interest. You can know with certainty which configurations have the most favorable properties. Another advantage of a minimalist model is that you don't have to be an expert in protein chemistry or molecular dynamics to play with it. A curious amateur can write a rudimentary program in a few days or weeks, and run it on commonly available machinery. Indeed, the simplified protein structures are so well suited to the needs of the amateur that I am tempted to call them amteins—they're not quite ready to turn pro yet. However, I have been persuaded to choose a name slightly less facetious, and so I shall call them prototeins. Foursquare FoldingThe specific model I've been toying with was devised 10 years ago by Ken A. Dill of the University of California at San Francisco, who has continued to explore it since then with the help of several colleagues. Almost all of my experiments merely replicate their earlier work. Dill's molecules would not be recognized as proteins by a biochemist (or by a ribosome, for that matter). They are radically simplified in three ways. First, whereas real proteins are constructed from 20 kinds of amino acids (which differ in size, shape, electric charge, affinity for water and other properties), the building blocks of prototeins come in just two flavors. Dill designates them H and P, for hydrophobic and polar; the H units repel water while the P units attract it. Second, the various forces acting between amino acids in proteins (electrostatic attractions and repulsions, hydrogen bonds, solvent interactions) are reduced in prototeins to a single rule: H's like to stick together. The P units in prototeins are inert, neither attracting nor repelling. Third, prototeins do their folding on a lattice, as if the molecules were laid out on graph paper. Think of the H's and P's as colored dots placed at the grid points of the lattice; the chemical bonds in the backbone of the prototein are lines drawn on the grid to connect the dots. Confining the molecules to a lattice is a major computational convenience. It keeps the number of configurations finite. If the chain could bend and twist in continuous space, there would be no clear way of counting the arrangements, and you could never be sure you had tried them all. Dill and others have explored several lattice geometries in both two and three dimensions. My own experiments all inhabit the two-dimensional square lattice, which is the simplest. Dots and lines on graph paper: That's really all there is to a prototein. Or else the model could be described in terms of colored beads, laid down on a board with a gridlike pattern of dimples to hold the beads in place. To build a sequence of amino acids, you string together H-beads and P-beads in whatever order you choose. To fold the molecule, you arrange the string of beads on the lattice board. The string is not allowed to stretch or break, and so successive beads in the sequence have to occupy nearest-neighbor sites on the lattice. No two beads can be piled up at the same site, and so the chain cannot cross itself. If two H-beads that are not adjacent within the linear sequence wind up on adjacent sites after the chain is folded, their attraction creates a cross-link, or contact, that helps to stabilize the molecule. Foldings that give rise to many such contacts are favored over those with few contacts. Simple and abstract the model surely is—so much so that you can't help wondering if the process of abstraction hasn't sucked all the life out of it. The squared-off, flattened molecules certainly don't look very biological. But the proof of the prototein is in the folding. Self-AvoidanceA program for studying prototeins has two main tasks to accomplish. The first chore is to generate all possible sequences of H's and P's. This part is easy; it's just binary counting. Any prototein sequence of length r can be mapped onto an r-bit binary number, simply by replacing each 1 in the binary representation of the number with an H, and each 0 with a P. The complete set of r-bit sequences is enumerated by counting from 0 to 2r–1. For example, in the case of r = 5 there are 32 sequences, starting with PPPPP, PPPPH and PPPHP, and continuing through HHHHH. The program's second task is to generate all possible foldings of each sequence. This is a little more challenging. A folding is modeled by a self-avoiding walk: a path through the lattice that visits no site more than once. The shortest self-avoiding walks are easy to analyze. On the square lattice there are exactly four self-avoiding walks one step long, namely the walks that move one site north, east, west or south of the origin. Each of these walks can be extended in three different ways to form two-step walks. The walk that begins with an eastward step can continue with a second step to the east, north or south; it cannot go west, because it would be retracing its own steps, and that is forbidden. Thus there are 4 x 3 = 12 walks of two steps each. The same kind of reasoning shows there are 36 three-step walks. But beyond this point the counting begins to get messy. Consider the three-step walk that goes first east, then north, then west. On the fourth step this walk cannot turn east, since that would constitute illegal backtracking. It also cannot go south, since it would thereby return to the origin—a site it has already visited. Hence there are only two available directions for this particular walk, whereas some other walks still have three options. It gets even worse: A walk can box itself in so that there are no legal moves, and the walk has to be abandoned. When I began experimenting with algorithms for self-avoiding walks, I found them so diverting that I thought I might never get back to the larger project of folding proteins. I could easily fill up an entire column with self-avoiding walks—and so that is what I have decided to do. I will make them the subject of a future column, and here give only a brief summary of how they fit into the world of prototeins. To survey all possible foldings of a prototein of r beads, you must generate all self-avoiding walks of r–1 steps. There is no shortcut for producing the complete set of walks; you have to enumerate them all. And each time you add a step, you have to check to make sure the destination site is not already occupied. There are tricks for speeding up the process, but none of them fundamentally change the nature of the algorithm. As the walks get longer, the effort of counting them grows exponentially; adding one step multiplies the number of walks by about 2.6. Through a prodigious feat of computing, A. R. Conway and A. J. Guttmann have counted all the self-avoiding walks of up to 51 steps (there are more than 1022), but for the amateur in self-avoidance the practical limit is probably between 20 and 30 steps. If your computer has enough memory, you can store a list of walks rather than regenerate them for each prototein sequence; this saves a great deal of time. Symmetries can reduce the number of walks you need to generate or store. For the purposes of molecular modeling, taking two steps east and one step north is no different from going two steps north and one step west; the paths are the same but for a 90-degree rotation. When all such symmetries are taken into account, the number of unique walks is cut to approximately 1/16th the total number. But there are still plenty of walks. At a length of 15 steps, 401,629 unique walks remain after all symmetries are eliminated. Given a procedure for generating binary H-P sequences and another procedure for generating self-avoiding walks, it is a simple matter to combine them. The idea is to produce all possible combinations of sequences and walks, folding up each sequence into the geometry defined by each walk. From this collection of folded molecules you can then gather statistical information—such as the average number of H-H contacts—or search for notably good foldings. High-Scoring MoleculesWhat makes for a good folding? In proteins the usual measure is the Gibbs free energy, a thermodynamic quantity that depends on both energy and entropy. If you could tug on the ends of a protein chain and straighten it out, the result would be a state of high energy and low entropy. The energy is high because amino acids that "want" to be close together are held at a distance; the entropy is low because the straight chain is a highly ordered configuration. When you let go, the chain springs back into a shape with lower energy and higher entropy, changes that translate into a lower value of the Gibbs free energy. The "native" state of a protein—the folding it adopts under natural conditions—is usually assumed to be the state with the lowest possible free energy. Prototeins can get along with a simpler folding criterion. Standard practice is to rank foldings simply by counting H-H contacts. It's more like keeping score than measuring energy. If the H's are viewed as analogues of hydrophobic amino acids, the scoring system reflects the tendency of hydrophobic groups to seek shelter from water. But the prototein model is so abstract that it doesn't really matter what kind of force is at play between the H's. Just say that H's are sticky, and it takes energy to pull them apart. One strategy for finding good folds, then, is to look for configurations that maximize the number of H-H contacts. A program to carry out the search runs through all the foldings of all the sequences of a given length, keeping only those foldings with the maximum number of contacts. How many contacts are possible in a folded prototein? A little doodling on graph paper shows that the highest possible ratio of contacts to H's is 7:6. Sequences that attain this limit are exceedingly rare. (I leave it as a puzzle for the reader to find the shortest such sequence, which I believe has 26 beads.) But proteins are not required to solve such mathematical puzzles. To find the stablest configurations of a given sequence, all you need do is find the foldings that have more H-H contacts than any other foldings of the same sequence, whether or not the number of contacts is the theoretical maximum. There is a shortcut for identifying these stable foldings. It begins with the sequence made up entirely of H's, which is rather like double-sided sticky tape that collapses on itself in a crumpled ball. If any sequence at all has a folding with a given number of H-H contacts, then that configuration must also be among the stablest foldings of the all-H sequence. In the all-H folding, however, some of the H's may not form contacts, and so they can be changed to P's without altering the score of the folding. By making all such substitutions, you recover the sequence with the minimum number of H's that can give rise to a given folding. Sequences with rigid, heavily cross-linked folds are fairly rare. Among chains with 21 beads the maximum number of H-H contacts is 12, and a chain must have at least 14 H's to reach this limit. There are only 80 sequences of 14 H's and 7 P's that produce 12 contacts, out of the universe of more than two million 21-bead sequences.
Figure 1 shows some of the 80 maximally cross-linked 21-bead prototeins, along with a few other foldings chosen at random. The two populations of molecules are very different. The randomly chosen configurations tend to be loose and floppy, and their average number of H-H contacts works out to less than 1. The highest-scoring folds, in contrast, are all very compact, with the chain either wound around itself in a spiral shape or folded into zigzags. A lifelike feature of the compact foldings is a tendency for the H's to congregate in the interior of the molecule, leaving the P's exposed on the surface. The model has no explicit rule favoring the formation of such a hydrophobic core; it happens automatically when you select foldings with numerous H-H contacts. In this connection, Dill points out that for short prototein chains a two-dimensional lattice model may be more realistic than a three-dimensional one. The reason is that the perimeter-to-area ratio of a short chain in two dimensions approximates the surface-to-volume ratio of a longer chain in three dimensions. Not all features of the high-scoring prototein foldings inspire confidence in the model's realism. For example, a disproportionate number of the best sequences have H's at both ends, and these molecules tend to fold up with their ends tucked into the hydrophobic core. The reason is easy to see: An H at the end of a chain can participate in three contacts, whereas interior H's can have no more than two. But the sticky-end effect is an artifact of the model; there is no comparable phenomenon in real proteins. Another peculiarity can be traced to the choice of a square lattice. Two H's on a square lattice can form a contact only if they are separated within the prototein sequence by an even number of intervening beads. As a result, every prototein can be divided into odd and even subsequences that do not interact. No such parity effect is seen in proteins. This failure of realism is unfortunate; on the other hand, the segregation of odd and even sublattices allows some very handy optimizations in a simulation program. Escaping Degeneracy
Are folds that maximize the number of H-H contacts the best folds for a prototein? Not necessarily. A high H-H score enhances a molecule's stability, which is certainly a useful property in a biopolymer, but there are other factors to consider as well. Stability implies that once a molecule is folded, it will probably stay folded. It's also important, however, that all molecules with the same sequence fold up to yield the same structure. The way to achieve such uniformity is to select sequences that have a unique best folding, even if that folding does not have the highest possible H-H score. A molecule with many equally good foldings is said to have a degenerate ground state. The all-P sequence is an obvious example: Every folding has an H-H score of zero. The all-H sequence is also degenerate. Obviously, any sequences with unique preferred foldings must be found between these extremes, but the existence of such sequences cannot be taken for granted. You can search for them by sorting all the foldings of a sequence into bins according to their H-H score; if the highest-scoring bin has a single occupant, that sequence has a unique best folding.
On the square lattice, uniquely folding sequences do exist for all but one of the chain lengths I was able to test. (The exception is length 5.) The longest chains I examined have 14 beads. Among the 16,384 sequences of this length, 955 have a unique folding. Within this subset, 96 foldings have seven H-H contacts, which is the maximum observed in 14-bead prototeins. The sequences in this elite subset, combining uniqueness with high stability, might be considered among the most lifelike prototeins. Low degeneracy and numerous contacts are not the only criteria for judging a prototein fold. Martin Karplus and Eugene Shakhnovich work with three-dimensional lattice models and employ a more realistic energy spectrum than the simple contact counting of the H-P scheme. Their findings highlight the importance of having a large energy gap between the best folding and the next-best one. They have also looked into the kinetics of folding, asking not just which configuration is stablest but also how long it takes a randomly wriggling molecule to find that conformation. Among 200 candidate sequences, 30 repeatedly discovered the state of minimum energy after no more than 50 million small random rearrangements. How Do Proteins Do It?
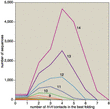
Although the prototein model is only a crude caricature of real protein folding, even this simplified simulation can be computationally taxing. For prototeins of length r, the number of sequences is 2r, and the number of foldings is approximately 2.6r–1; the effort needed to solve a model is proportional to the product of these numbers. That product grows steeply. The five-bead model can be solved by hand, and a commodity computer disposes of the 10-bead model in seconds. But a chain of 15 beads combines 32,768 sequences with 148,752 folds, for a total of almost 5 billion cases. At 20 beads, the product of sequences and folds is over 20 trillion, which is way beyond the limit of this amateur's patience (and lifespan). Attacking these models with brute-force computations could turn out to be comically stupid. Maybe there's some clever algorithm waiting to be discovered that will make folding easy. Maybe, but not likely. In the past few months two groups have proved that models much like the one described here belong in the class of hard problems known as NP-complete. Bonnie Berger and Tom Leighton give a proof for the three-dimensional H-P model. The two-dimensional case is proved in a quite different way by Pierluigi Crescenzi, Deborah Goldman, Christos Papadimitriou, Antonio Piccolboni and Mihalis Yannakakis. Showing that a problem is NP-complete doesn't actually prove it is hard; NP-completeness merely certifies that the problem is as hard as a bunch of others, and a method for efficiently solving any one of the problems could be adapted to all the rest. Some miraculous algorithm could sweep the whole class of problems away. But don't hold your breath. Which brings us back to Levinthal's question: If protein folding is so hard, how do proteins do it? There are three kinds of answers. One possibility is that protein molecules are capable of mathematical wizardry beyond the reach of conventional computers. This would stand the NP-completeness result on its head; instead of proving that protein folding is hard, it would show that everything else is easy. You could encode an instance of any NP-complete problem in a synthetic sequence of amino acids, then let the protein fold itself up; from the folded configuration you could read out the solution to the original problem. Another answer is that proteins, contrary to their reputation, do not always fold efficiently and spontaneously. Some of them need help, in the form of "chaperone" molecules. Some may fold erroneously and be recycled by proteolytic enzymes. And it's possible that the native state of some proteins is not in fact the state of lowest free energy. A biological molecule doesn't have to be absolutely stable; it only has to last long enough to do its job. Perhaps the appropriate model of protein folding is not an exhaustive search for the best conformation but an approximation algorithm that is guaranteed to quickly find a good folding. William E. Hart and Sorin Istrail have published just such an algorithm for the H-P model. The third option is that proteins do quickly find the best among all possible foldings, but only because they have evolved to exhibit precisely this property. In other words, the only amino acid sequences that survive under natural selection are those that happen to fold rapidly. Sequences that fold hierarchically could fit this description: If small sections of the chain condense independently into secondary structures such as helices, which then aggregate without further internal rearrangement, the combinatorial monster might be tamed. Although this mechanism cannot explain everything about protein folding—indeed, Dill argues that secondary structures are a consequence of compact folding rather than a cause—it certainly helps. In any case, the idea that any arbitrary amino acid sequence would fold efficiently is surely overoptimistic—which nixes the fantasy of a protein-folding computer for NP-complete problems. But the subset of rapidly folding sequences remains poorly understood. Computational models, even simplistic ones, offer a means of probing it. In this connection it is worth noting that nature itself has hardly begun to explore the full space of amino acid sequences. All the proteins in all the organisms that ever lived on the earth could not sample more than an utterly negligible fraction of the 20100 or so possible sequences. Thus a computation something like the ones carried out in the H-P model is running at this moment, all over the planet, in the big green computer.
© Brian Hayes |
Bibliography
- Berger, Bonnie, and Tom Leighton. 1998. Protein folding in the hydrophobic-hydrophilic (HP) model is NP-complete. Journal of Computational Biology, 5(1):27–40.
- Conway, A. R., and A. J. Guttmann. 1996. Square lattice self-avoiding walks and corrections to scaling. Physical Review Letters 77:5284–5287. [CrossRef]
- Crescenzi, Pierluigi, Deborah Goldman, Christos Papadimitriou, Antonio Piccolboni and Mihalis Yannakakis. 1998. On the complexity of protein folding. In Proceedings of the Second International Conference on Computational Molecular Biology (RECOMB), New York, NY, March 22?25, 1998. http://http.cs.berkeley.edu/~christos/hp.ps
- Dill, Ken A., Sarina Bromberg, Kaizhi Yue, Klaus M. Fiebig, David P. Yee, Paul D. Thomas and Hue Sun Chan. 1995. Principles of protein folding?A perspective from simple exact models. Protein Science 4:561–602.
- Fraenkel, Aviezri S. 1993. Complexity of protein folding. Bulletin of Mathematical Biology 55:1199–1210.
- Hart, William E., and Sorin Istrail. 1995. Fast protein folding in the hydrophobic-hydrophilic model within three-eighths of optimal. Proceedings of the 27th Annual ACM Symposium on the Theory of Computing, May 29?June 1, 1995, Las Vegas, Nevada, pp. 157–168.
- Lau, K. F., and Ken A. Dill. 1989. A lattice statistical mechanics model of the conformational and sequence spaces of proteins. Macromolecules 22:3986–3997.
- Levinthal, Cyrus (notes by A. Rawitch). 1969. How to fold graciously. In M?ssbauer Spectroscopy in Biological Systems, edited by P. Debrunner, J. C. M. Tsibris, and E. M?nck. Proceedings of a meeting held at Allerton House, March 17 and 18, 1969, Monticello, Illinois. Urbana, Ill.: University of Illinois Press.
- Merz, Kenneth M., Jr., and Scott M. Le Grand (editors). 1994. The Protein Folding Problem and Tertiary Structure Prediction. Boston: Birkh?user.
- Sali, Andrej, Eugene Shakhnovich and Martin Karplus. 1994. Kinetics of protein folding: A lattice model study of the requirements for folding to the native state. Journal of Molecular Biology 235:1614–1636.
- Yue, Kaizhi, and Ken A. Dill. 1995. Forces of tertiary structural organization of globular proteins. Proceedings of the National Academy of Sciences of the U.S.A. 92:146–150.
- Yue, Kaizhi, Klaus M. Fiebig, Paul D. Thomas, Hue Sun Chan, Eugene I. Shakhnovich and Ken A. Dill. 1995. A test of lattice protein folding algorithms. Proceedings of the National Academy of Sciences of the U.S.A. 92:325–329.
- Yue, Kaizhi, and Ken A. Dill. 1996. Folding proteins with a simple energy function and extensive conformational searching. Protein Science 5:254–261
|
Of Possible Interest
Column: Computing Comes to Life
Column: Computational Creationism
Column: The World According to Wolfram
Column: The Weatherman
Column: Follow the Money
| ||
| Related Sigma Xi Links |