Research Summary
Research in our laboratory is focused on the problem of polarity, using the Xenopus oocyte as a model system. Our long-term goal is to understand the molecular mechanism by which informational molecules, such as mRNA and protein, are localized to specified regions of the cell cytoplasm.
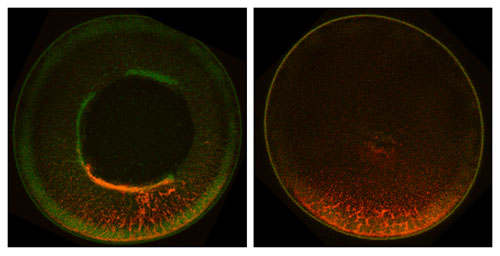
We have studied the localization of Vg1 mRNA in the frog oocyte as a model to gain mechanistic insight into how RNA molecules can be targeted to specific regions of the cell cytoplasm to generate spatially restricted gene expression. This process has emerged as a widespread mechanism for generating cell polarity in somatic cells and can provide the basis for patterning during embryonic development. In vertebrates, a paradigm of this phenomenon is localization of Vg1 RNA within the Xenopus oocyte, a process directed by recognition of an RNA localization element within the Vg1 3' UTR. To probe the biochemical mechanisms underlying the localization process, we have studied the RNA-protein interactions leading to formation of a Vg1 RNP transport complex. Based on our results, we have proposed a model for cytoplasmic RNA localization in which a core localization RNP is initially assembled in the nucleus, where factors specific nuclear RNA binding proteins associate with the RNA. Upon transport from the nucleus to the cytoplasm, the RNP complex is remodeled and additional transport factors are recruited. After motor-directed transport to the vegetal cortex, RNP complex is subsequently anchored at the vegetal cortex. Our work has uncovered distinct nuclear and cytoplasmic steps in the localization pathway, and our ongoing and future studies are designed to provide new molecular insights into the RNA localization pathway, linking the earliest steps in the pathway with the motors that accomplish the transport process.
