- Computational Models of SNPs and Disease Associations
- CELLARIUM: An Integrated Framework for Genome Browsers and Bioinformatics Workflows
- Medical Bioinformatics and Beyond
- Algorithmic Theory of Protein Folding and Misfolding
- Statistical Mechanics and Computational Complexity
- Education and the Classroom
This work was funded by the National Science Foundation

Project Resources
- Overview
- Collaborators
- Publications
- Related Software
- Presentations
- Posters
Albert Einstein (1953)
Overview



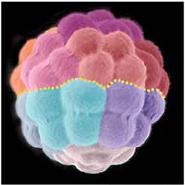
We report the sequence and analysis of the 814-megabase genome of the sea urchin Strongylocentrotus purpuratus, a model for developmental and systems biology. The sequencing strategy combined whole-genome shotgun and bacterial artificial chromosome (BAC) sequences. This use of BAC clones, aided by a pooling strategy, overcame difficulties associated with high heterozygosity of the genome. The genome encodes about 23,300 genes, including many previously thought to be vertebrate innovations or known only outside the deuterostomes. This echinoderm genome provides an evolutionary outgroup for the chordates and yields insights into the evolution of deuterostomes.
The sea urchin Strongylocentrotus purpuratus is a model organism for study of the genomic control circuitry underlying embryonic development. We examined the complete repertoire of genes expressed in the S. purpuratus embryo, up to late gastrula stage, by means of high-resolution custom tiling arrays covering the whole genome. We detected complete spliced structures even for genes known to be expressed at low levels in only a few cells. At least 11,000 to 12,000 genes are used in embryogenesis. These include most of the genes encoding transcription factors and signaling proteins, as well as some classes of general cytoskeletal and metabolic proteins, but only a minor fraction of genes encoding immune functions and sensory receptors. Thousands of small asymmetric transcripts of unknown function were also detected in intergenic regions throughout the genome. The tiling array data were used to correct and authenticate several thousand gene models during the genome annotation process.
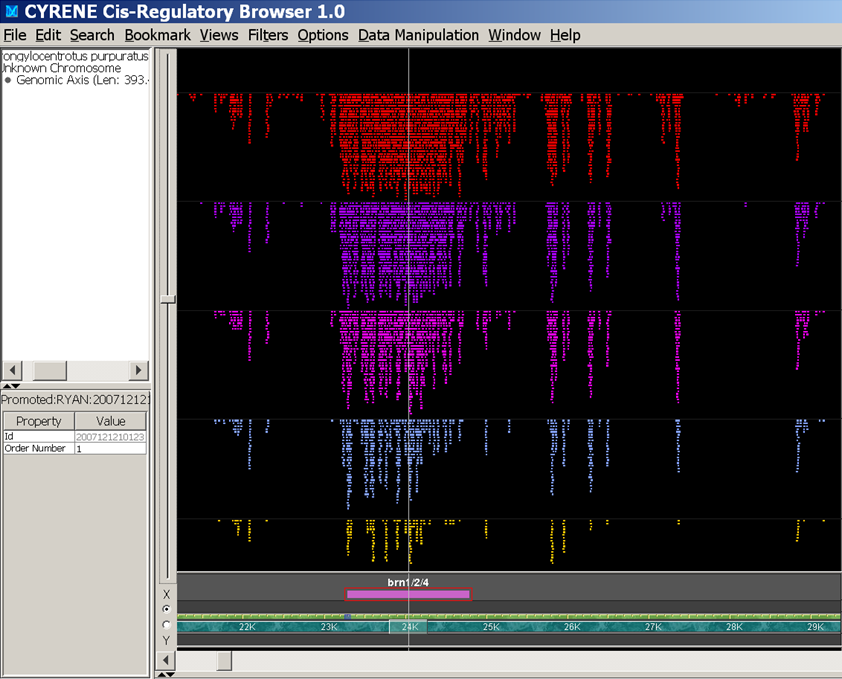
cis-regulatory modules that control developmental gene expression process the regulatory inputs provided by the transcription factors for which they contain specific target sites. A prominent class of cis-regulatory processing functions can be modeled as logic operations. Many of these are combinatorial because they are mediated by multiple sites, although others are unitary. In this work, we illustrate the repertoire of cis-regulatory logic operations, as an approach toward a functional interpretation of the genomic regulatory code.
The definitive feature of the many thousand cis-regulatory control modules in an animal genome is their information processing capability. These modules are “wired” together in large networks that control major processes such as development; they constitute “genomic computers.” Each control module receives multiple inputs in the form of the incident transcription factors which bind to them. The functions they execute upon these inputs can be reduced to basic AND, OR and NOT logic functions, which are also the unit logic functions of electronic computers. Here we consider the operating principles of the genomic computer, the product of evolution, in comparison to those of electronic computers. For example, in the genomic computer intra-machine communication occurs by means of diffusion (of transcription factors), while in electronic computers it occurs by electron transit along pre-organized wires. There follow fundamental differences in design principle in respect to the meaning of time, speed, multiplicity of processors, memory, robustness of computation and hardware and software. The genomic computer controls spatial gene expression in the development of the body plan, and its appearance in remote evolutionary time must be considered to have been a founding requirement for animal grade life.
Gene expression is controlled by interactions between trans-regulatory factors andcis-regulatory DNA sequences, and these interactions constitute the essential functional linkages of gene regulatory networks (GRNs). Validation of GRN models requires experimental cisregulatory tests of predicted linkages to authenticate their identities and proposed functions. However, cis-regulatory analysis is, at present, at a severe bottleneck in genomic system biology because of the demanding experimental methodologies currently in use for discovering cis-regulatory modules (CRMs), in the genome, and for measuring their activities. Here we demonstrate a high-throughput approach to both discovery and quantitative characterization of CRMs. The unique aspect is use of DNA sequence tags to “barcode” CRM expression constructs, which can then be mixed, injected together into sea urchin eggs, and subsequently deconvolved. This method has increased the rate of cis-regulatory analysis by >100-fold compared with conventional one-by-one reporter assays. The utility of the DNA-tag reporters was demonstrated by the rapid discovery of 81 active CRMs from 37 previously unexplored sea urchin genes. We then obtained simultaneous high-resolution temporal characterization of the regulatory activities of more than 80 CRMs.On average 2–3 CRMs were discovered per gene. Comparison of endogenous gene expression profiles with those of the CRMs recovered from each gene showed that, for most cases, at least one CRM is active in each phase of endogenous expression, suggesting that CRM recovery was comprehensive. This approach will qualitatively alter the practice of GRN construction as well as validation, and will impact many additional areas of regulatory system biology
Current project members: Ryan Tarpine
Project
alumni: Rohan Maddamsetti, Sanjay Trehan, and David Moskowitz
Collaborators
- Norman Chandler Professor of Cell Biology Eric Davidson, California Institute of Technology
- Joel Smith, Ph.D., Marine Biological Laboratory, Woods Hole
- Russel Turner, Ph.D.
- Professor Gary Wessel, Brown University
Publications
| 2012 | |
| [8] | Hagit Shatkay, Ramya Narayanaswamy, Santosh Nagaral, Na Harrington, Dorothea Blostein, Ryan Tarpine, Kyle Schutter, Rohith Mv, Gowri Somanath, Sorin Istrail, Chandra Kambahmettu, "OCR-based Image Features for Biomedical Image and Article Classification: Identifying Documents Relevant to Cis-Regulatory Elements", In ACM BCB, 2012. [bib] [pdf] |
| 2010 | |
| [7] | Jongmin Nam, Ping Dong, Ryan Tarpine, Sorin Istrail, Eric H. Davidson, "Functional cis-regulatory genomics for systems biology", In Proceedings of the National Academy of Sciences, vol. 107, no. 8, pp. 3930-3935, 2010. [bib] [pdf] [doi] |
| [6] | Sorin Istrail, Ryan Tarpine, Kyle Schutter, Derek Aguiar, "Practical Computational Methods for Regulatory Genomics: A cisGRN-Lexicon and cisGRN-Browser for Gene Regulatory Networks", Chapter in Computational Biology of Transcription Factor Binding, Humana Press, vol. 674, pp. 369-399, 2010. [bib] [pdf] [doi] |
| 2009 | |
| [5] | Ryan Tarpine, Sorin Istrail, "On the Concept of Cis-Regulatory Information: From Sequence Motifs to Logic Functions", Chapter in Algorithmic Bioprocesses, Springer-Verlag, pp. 731-742, 2009. [bib] [pdf] |
| 2007 | |
| [4] | Sorin Istrail, Smadar Ben-Tabou De-Leon, Eric H. Davidson, "The regulatory genome and the computer", In Developmental Biology, vol. 310, no. 2, pp. 187-195, 2007. [bib] [pdf] [doi] |
| 2006 | |
| [3] | Manoj P. Samanta, Waraporn Tongprasit, Sorin Istrail, R. Andrew Cameron, Qiang Tu, Eric H. Davidson, Viktor Stolc, "The Transcriptome of the Sea Urchin Embryo", In Science, vol. 314, no. 5801, pp. 960-962, 2006. [bib] [pdf] [doi] |
| [2] | Sea Urchin Genome Sequencing Consortium, Erica Sodergren, George M. Weinstock, Eric H Davidson, R. Andrew Cameron, Richard A. Gibbs, Robert C. Angerer, Lynne M. Angerer, Maria Ina Arnone, David R. Burgess, Robert D. Burke, James A. Coffman, Michael Dean, Maurice R. Elphick, Charles A. Ettensohn, Kathy R. Foltz, Amro Hamdoun, Richard O. Hynes, William H. Klein, William Marzluff, David R. McClay, Robert L. Morris, Arcady Mushegian, Jonathan P. Rast, L. Courtney Smith, Michael C. Thorndyke, Victor D. Vacquier, Gary M. Wessel, Greg Wray, Lan Zhang, Christine G. Elsik, Olga Ermolaeva, Wratko Hlavina, Gretchen Hofmann, Paul Kitts, Melissa J. Landrum, Aaron J. Mackey, Donna Maglott, Georgia Panopoulou, Albert J. Poustka, Kim Pruitt, Victor Sapojnikov, Xingzhi Song, Alexandre Souvorov, Victor Solovyev, Zheng Wei, Charles A. Whittaker, Kim Worley, K. James Durbin, Yufeng Shen, Olivier Fedrigo, David Garfield, Ralph Haygood, Alexander Primus, Rahul Satija, Tonya Severson, Manuel L. Gonzalez-Garay, Andrew R. Jackson, Aleksandar Milosavljevic, Mark Tong, Christopher E. Killian, Brian T. Livingston, Fred H. Wilt, Nikki Adams, Robert Bell??, Seth Carbonneau, Rocky Cheung, Patrick Cormier, Bertrand Cosson, Jenifer Croce, Antonio Fernandez-Guerra, Anne-Marie Genevi??re, Manisha Goel, Hemant Kelkar, Julia Morales, Odile Mulner-Lorillon, Anthony J. Robertson, Jared V. Goldstone, Bryan Cole, David Epel, Bert Gold, Mark E. Hahn, Meredith Howard-Ashby, Mark Scally, John J. Stegeman, Erin L. Allgood, Jonah Cool, Kyle M. Judkins, Shawn S. McCafferty, Ashlan M. Musante, Robert A. Obar, Amanda P. Rawson, Blair J. Rossetti, Ian R. Gibbons, Matthew P. Hoffman, Andrew Leone, Sorin Istrail, Stefan C. Materna, Manoj P. Samanta, Viktor et al. Stolc, "The Genome of the Sea Urchin Strongylocentrotus purpuratus", In Science, vol. 314, no. 5801, pp. 941-952, 2006. [bib] [pdf] [doi] |
| 2005 | |
| [1] | Sorin Istrail, Eric Davidson, "Logic functions of the genomic cis-regulatory code", In Proceedings of the National Academy of Sciences, vol. 102, no. 14, pp. 4954-4959, 2005. [bib] [pdf] |
