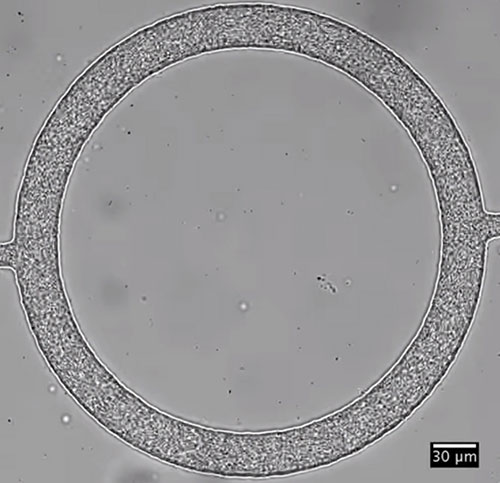
Bacteria clustered together in tiny racetracks exhibit complex collective dynamics. New research shows that those dynamics emerge due to collisions and fluid flow. H. Wioland/E. Lushi/R.E. Goldstein
PROVIDENCE, R.I. [Brown University] — As the world prepares to watch the Summer Olympics’ track and field events in Rio, it will come as no surprise that the runners in each race travel in the same direction around the track. But new research shows that if those runners were bacteria, the dynamics on the track would be a bit more complex — and more than a little puzzling.
According to work by an international group of researchers, when bacteria are confined in millimeter-sized, fluid-filled oval “racetracks,” they quickly organize themselves into a clear collective motion. But instead of all traveling in one direction, individual bacteria along the outside of the track go one way, while those in the middle go the other.
The aim of the work was to better understand how bacterial colonies spread though tiny water-filled cracks and crevices they inhabit in natural settings like rock, soil or body tissue. And thanks to these experiments and a set of computer models, the researchers believe they’ve figured out how these complex dynamics emerge in colonies of rather unsophisticated creatures.
The study, by researchers from Brown University and the University of Cambridge, is published in the New Journal of Physics.
Nature is full of collective patterns that emerge from the behavior of individual organisms. Flocks of starlings, for example, can often be seen creating intricate, flowing patterns in the sky. Bacteria are known to exhibit these kinds of seemingly coordinated patterns as well.
“It’s not at all obvious how they do this,” said Enkeleida Lushi, a researcher at Brown University’s School of Engineering and one of the study’s co-authors. “Bacteria don’t have leaders to follow, they have limited sensory systems, and they’re not very smart. They can’t really make decisions about where to go, so it is all down to mechanical interactions between themselves and their surroundings.”
For the past several years, Lushi and her colleagues have been working to understand how collective patterns in bacterial colonies form. For this latest research, they wanted to design an experiment that would mimic the tiny channels in which bacteria thrive in nature. Their solution was to make tiny racetracks from clear plastic, so they could observe the behavior of Bacillus subtilis bacteria under a microscope.
The experiments showed that the chaotic motion of all those individual swimmers quickly organizes itself into a collective motion, with bacteria in the middle of the track going in the opposite direction from those on the outer edges.
In order to understand what was driving that pattern, Lushi developed a computer model that captured two critical parameters that affect how the bacteria move in a densely packed colony. The first parameter involves the collisions that occur as bacteria try to swim in close quarters. The model showed that, as bacteria bang into each other, the collisions tend to orient individuals in the same direction.
As for the bi-directional motion — with the middle swimmers moving in opposite direction from the ones at the edges — that is a function of fluid flow, the research showed.
Bacteria propel by pushing against the tiny fluid corkscrew-like appendages called flagella. That pushing creates a fluid flow moving opposite the direction of the swimming. The bacteria along the edges of the track are closely aligned by the track’s outer surface. They swim at an angle against that outer surface, which causes the backflow they create to be aimed toward the middle of the track. The swimmers in the middle have to fight the currents generated on either side of them. Eventually, the flow becomes more than they can handle.
“Even though they’re trying to go in the same direction as the ones on the boundaries, they’re being carried backward by the fluid,” Lushi said.
The results, Lushi says, underscore the importance of fluid flows in explaining collective dynamics. Despite the fact that most bacteria thrive and have evolved in fluid environments, the effects of fluid flows are often overlooked in explaining their behavior.
Eventually, Lushi and her colleagues are hopeful that research like this might deepen our understanding of how bacteria spread in their natural environment. It could also aid in the development of medical devices with surfaces and architectures that can manipulate bacterial movements and spread.
Those applications, however, remain a bit further down the road, Lushi said. “At this point, we’re still taking baby steps to learn about the bacterial interactions and behaviors.”
Lushi’s co-authors on the paper were Hugo Wioland and Raymond Goldstein from the University of Cambridge. The work was supported by the European Research Council (grant 247333) and the National Science Foundation (CBET-1544196)