PROVIDENCE, R.I. [Brown University] — Computer models developed by Brown University mathematicians show new details of what happens inside a red blood cell affected by sickle cell disease. The researchers said they hope their models, described in an article in the Biophysical Journal, will help in assessing drug strategies to combat the genetic blood disorder, which affects millions of people worldwide.
Sickle cell disease affects hemoglobin, molecules within red blood cells responsible for transporting oxygen. In normal red blood cells, hemoglobin is dispersed evenly throughout the cell. In sickle red blood cells, mutated hemoglobin can polymerize when deprived of oxygen, assembling themselves into long polymer fibers that push against the membranes of the cells, forcing them out of shape. The stiff, ill-shaped cells can become lodged in small capillaries throughout the body, leading to painful episodes known as sickle cell crisis.
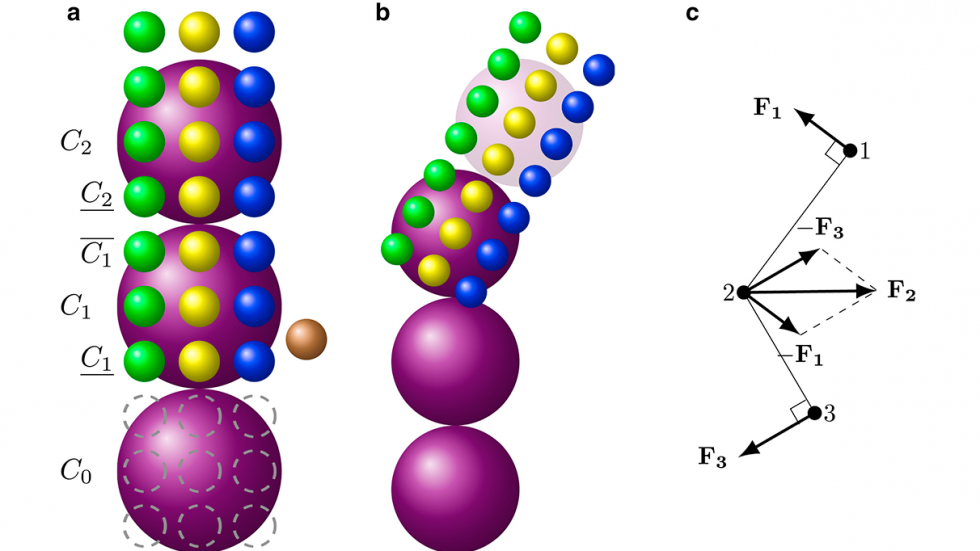
“The goal of our work is to model both how these sickle hemoglobin fibers form as well as the mechanical properties of those fibers,” said Lu Lu, a Ph.D. student in Brown Division of Applied Mathematics and the study’s lead author. “There had been separate models for each of these things individually developed by us, but this brings those together into one comprehensive model.”
The model uses detailed biomechanical data on how sickle hemoglobin molecules behave and bind with each other to simulate the assembly of a polymer fiber. Prior to this work, the problem had been that as the fiber grows, so does the amount of data the model must crunch. Modeling an entire polymer fiber at cellular scale using the details of each molecule was simply too computationally expensive.
“Even the world’s fastest supercomputers wouldn’t be able to handle it,” said George Karniadakis, professor of applied math at Brown and the paper’s senior author. “There’s just too much happening and no way to capture it all computationally. That’s what we were able to overcome with this work.”
The researchers’ solution was to apply what they call a mesoscopic adaptive resolution scheme or MARS. The MARS model calculates the detailed dynamics of each individual hemoglobin molecule only at the each end of polymer fibers, where new molecules are being recruited into the fiber. Once four layers of a fiber have been established, the model automatically dials back the resolution at which it represents that section. The model retains the important information about how the fiber behaves mechanically, but glosses over the fine details of each constituent molecule.
“By eliminating the fine details where we don’t need them, we develop a model that can simulate this whole process and its effects on a red blood cell,” Karniadakis said.
Using the new MARS simulations, the researchers were able to show how different configurations of growing polymer fibers are able to produce cells with different shapes. Though the disease gets its name because it causes many red blood cells take on a sickle-like shape, there are actually a variety of abnormal cell shapes present. This new modeling approach showed new details about how different fiber structures inside the cell produce different cell shapes.
“We are able to produce a polymerization profile for each of the cell types associated with the disease,” Karniadakis said. “Now the goal is to use these models to look for ways of preventing the disease onset.”
There are only two drugs on the market that has been approved by the FDA for treating sickle cell, Karniadakis says. One of them, called hydroxyurea, is thought to work by boosting the amount of fetal hemoglobin — the kind of hemoglobin that babies are born with — in a patient’s blood. Fetal hemoglobin is resistant to polymerization and, when present in sufficient quantity, is thought to disrupt the polymerization of sickle cell hemoglobin.
Using these new models, Karniadakis and his colleagues can now run simulations that include fetal hemoglobin. Those simulations could help to confirm that fetal hemoglobin does indeed disrupt polymerization, as well as help to establish how much fetal hemoglobin is necessary. That could help in establishing better dosage guidelines or in developing new and more effective drugs, the researchers say.
“The models give us a way to do preliminary testing on new approaches to stopping this disease,” Karniadakis said. “Now that we can simulate the entire polymerization process, we think the models will be much more useful.”
Lu and Karniadakis’ co-authors on the paper were He Li, Xin Bian and Xuejin Li, all from Brown’s Division of Applied Mathematics. The work was supported by the National Institutes of Health (U01HL114476). Computer time and other resources were provided under grants from the Department of Energy (DE-AC02-06CH11357, DE-AC05-00OR22725).