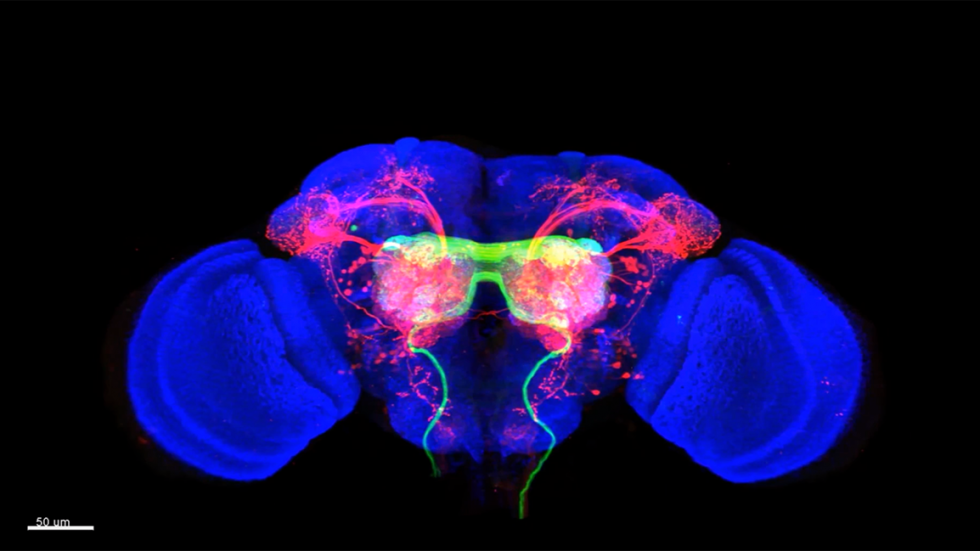
PROVIDENCE, R.I. [Brown University] — Finding out which neurons are connected with which others, and how they act together, is a huge challenge in neuroscience, and it’s crucial for understanding how brain circuits give rise to perception, motion, memory, and behavior. A new Brown University-developed technology called “trans-Tango" allows scientists to exploit the connections between pairs of neurons to make such discoveries in neuroscience. In a new study in Neuron, they used trans-Tango to illuminate connected neurons in fruit flies, revealing previously unmapped gustatory circuits that link the taste-sensing organs to brain regions known to govern feeding behavior and memory.
The technology is widely applicable, the researchers say, because trans-Tango doesn’t depend on the neurotransmitters involved in a neural connection or on the types of neurons that are connected. As long as two neurons join at a synapse, trans-Tango allows scientists to label the cells connected to a starter neuron, experiments in the paper show.
Moreover, because trans-Tango works by instigating the expression of genes in connected pairs of neurons, it also has the potential to enable scientists to control circuit functions, said senior and corresponding author Gilad Barnea, an associate professor of neuroscience at Brown who began looking for a precise, reliable and general way to visualize neural connections two decades ago. The application of trans-Tango that his team demonstrates in the new study is circuit tracing, but manipulations such as activating or shutting off connected neurons could become possible, too.
“trans-Tango provides genetic accessibility in the context of connectivity,” Barnea said. “Our technique allows you to access the neurons that interact with the particular ‘starter’ cell you target. It therefore expands the use of molecular genetic techniques beyond the cell for which you have a marker to the ones it ‘talks’ to.”
The team, which includes postdoctoral fellows, graduate students, research assistants and undergraduates, is now working on developing a host of other applications of trans-Tango. These include using the system to manipulate behavior, developing the equivalent technique in mice, and making it work in reverse so that it employs incoming connections from other neurons just like it does outgoing connections. That’s according to Mustafa Talay, a postdoctoral fellow who earned his Ph.D. in Barnea’s lab and is co-lead author with Ethan Richman, a former undergraduate at Brown who is now a graduate student at Stanford.
In addition, the Barnea lab is collaborating on adapting the technology to study how cancer spreads.
How it works
trans-Tango works by genetically introducing an artificial signaling pathway into every neuron in the fly. The pathway acts like a switch in the neurons that can be thrown by exposure to a triggering protein. To operate trans-Tango, scientists genetically engineer the neurons of interest (starter neurons) to present this triggering protein on their synapses together with a protein that lights up the starter neurons in green. Expression of the trigger protein at the synapse causes connected neurons to light up in red, revealing the full extent of the connected neurons in the fly’s nervous system.
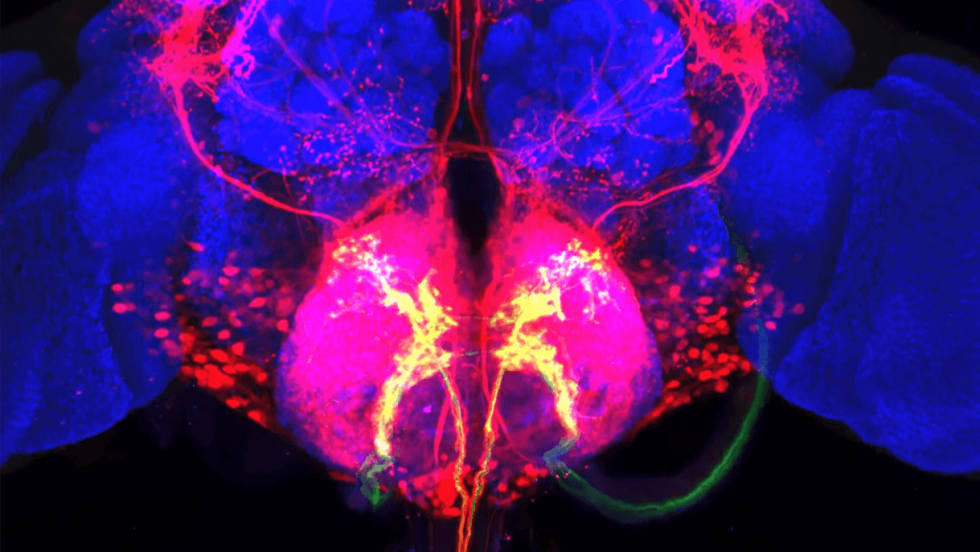
In the gustatory system, for example, the team lit up connections extending all the way from peripheral taste-sensing starter neurons to connected neurons that projected into a brain region known to control feeding behavior as well as to other regions thought to regulate memory.
By design, the system stops after just one stage of connectivity because if it continued endlessly, it would eventually light up the whole nervous system, Talay said. After all, each neuron usually connects to many others, not just one or a few, and ultimately they are pretty much all connected.
But the system is compatible with other cell imaging and targeting methods that can narrow down the number of connected neurons that respond to trans-Tango. In the new study, for example, the team combined trans-Tango with such techniques to specifically highlight individual connected neurons.
“When we probe a circuit we have no idea about, we can first just use trans-Tango and see the totality of all the connections of a neuron,” Talay said. “After that, if we want to characterize a circuit in more detail, we can combine trans-Tango with other methods to basically dissect that circuit.”
In many cases, revealing the full expanse that two connected neurons cover in a circuit can present deeply meaningful insights for neuroscientists. Not only did the team find novel connections in the gustatory circuitry of flies, but also they showed the different projections that various neurons in the olfactory system make, potentially clarifying how they carry out their distinct roles in connecting smell and behavior. Their experiments also highlighted connections that were already well known in the olfactory system, validating that the connections trans-Tango highlights are real.
The technology’s triggering protein is not naturally found in the fly, and it doesn’t leave the neurons or the synapse. For this reason, the scientists said, the illumination that arises as a result of trans-Tango reveals cells that truly “talk” to each other rather than neighboring but irrelevant cells.
How it was developed
Barnea has sought to perform exactly this kind of circuit mapping since he joined the lab of Columbia University Professor Richard Axel as a postdoctoral researcher in 1996. They were studying the olfactory system, and Barnea wanted to map the olfactory circuits in the rodent brain.
Tracing the connections of neurons within circuits in the brain is a fundamental but very difficult problem for neuroscientists. In all, the nervous systems of different organisms may involve many millions or billions of neurons with connections reaching into the trillions. It’s a lot to sort through.
There are several other methods for mapping circuits, but they all suffer from drawbacks. Some are too noisy. Some are too expensive and laborious. Some are too specific to a tiny subset of connections or neurons. Some only reveal the synapses but not the full length of the cells that connect there. Some won’t work in a living organism. Barnea wanted to generate a system for circuit mapping that would be general, precise, simple to use and that would work in an organism rather than in extracted tissue.
At Columbia, Barnea developed Tango, a method for studying cellular receptors that is the basis for the synthetic signaling pathway in trans-Tango. When he came to Brown in 2007, he continued this work and took on other projects. Barnea’s lab was not set for fly work, so its first fly incubator was an old egg incubator borrowed from biology professor Gary Wessel. The trans-Tango project was first supported by the Pew Charitable Trusts, then by the National Institutes of Health’s EUREKA program and subsequently by more conventional grants. The project also gained internal funding through the Innovation Award from the Brown Institute for Brain Science and Research Seed and Salomon awards from Brown’s Office of the Vice President for Research.
A key feature of trans-Tango is that it employs the human hormone glucagon as the trigger that switches the synthetic pathway on. Glucagon is engineered to localize to the synapse, and it is tethered in order to prevent it from diffusing away. Barnea credits the inspiration to use that form of glucagon to co-author John Szymanski, a former undergraduate student in his lab who is now a graduate student at Columbia. Szymanski first heard about the engineered form of glucagon at a party, Barnea said.
In 2011, Barnea met Talay while visiting Boğaziçi University in Turkey, where Talay was a master’s student. Talay was also thinking about ways to trace neural circuits and he had crucial experience working in flies, where progress could be faster than in mice.
Richman was interested in synthetic biology so he joined the Barnea lab to advance the development of the tracing technique. Talay and Richman led the charge to develop trans-Tango and make it work in flies, continually refining it with the help of several lab mates. This collaboration continued even after Richman graduated in 2013, when he decided to delay going to Stanford to see the project through.
“I remember very clearly the excitement of seeing the first images appear indicating a functioning technique, and the pleasure of discussing those results with Gilad,” Richman said. “That happened in January, and in the subsequent spring I had gotten accepted to graduate school and was slated to start the next fall. By the summer, Mustafa and I had made progress optimizing the technique, and the excitement in the lab was building. Having spent so long getting the technique to work, I was tantalized by the opportunity to put it into action.”
It was indeed a long time coming. Barnea points out that one of the paper’s co-authors, former undergraduate student Cambria Chou-Freed, is younger than the original idea he envisioned 21 years ago. In all, five of the paper’s authors were undergraduates in the lab, and all stayed in the lab after graduating to continue to work on this project.
“Everyone on the list of authors contributed something unique to the success of this project,” Barnea said. “This was driven by individuals who were committed and obsessed with it, but it was also very nice teamwork.”
The paper’s other authors are Nathaniel Snell, Griffin Hartmann, John Fisher, Altar Sorkaç, Juan Santoyo, Nived Nair and Mark Johnson.