PROVIDENCE, R.I.[Brown University] — Scientists have captured the first “snapshot” of two proteins involved in delivering a bacterial stress-response master regulator to the cell’s recycling machinery.
The Brown University-led team found that RssB — a protein that specifically recognizes the master regulator and delivers it to the recycling machinery somewhat like a recycling truck — forms a compact structure with a factor that inhibits RssB activity. The inhibition factor, called IraD, is triggered by DNA damage, one of many stresses the master regulator helps bacteria survive by turning on important genes.
The stress-response master regulator is critical for many bacteria — including those that cause human disease such as E. coli and Salmonella — to survive nutrient deprivation and other stressful situations such as those encountered during the process of infecting a new host. It is also important for the development and growth of biofilms, which are often resistant to antibiotics and grow on medical devices such as catheters and artificial heart valves.
“Before this study, people didn’t really understand how IraD acts to inhibit RssB,” said Alexandra Deaconescu, an assistant professor of molecular biology, cell biology and biochemistry at Brown and corresponding author for the study. “We were able to determine the structure of IraD bound to RssB and figure out how it works. Understanding how bacteria control the amount of stress-response master regulator could lead to the development of antimicrobials or even agents to halt the evolution of antibiotic resistance.”
It’s important for bacteria trying to establish an infection or growing in a stressful environment to be able to fine-tune the amount of the stress-response master regulator they have, as the regulator controls a large percentage of E. coli genes, which may need to be turned on under specific conditions, Deaconescu said. Recycling the regulator is wasteful of the chemical energy used to produce the regulator, but is a faster method of control than turning its production on or off, making it suitable for adapting to ever-changing environments. In fact, bacteria control both the production and recycling of the master regulator to adapt and thrive.
The structure of IraD bound to RssB and findings were published on Thursday, April 11, in the journal Genes and Development.
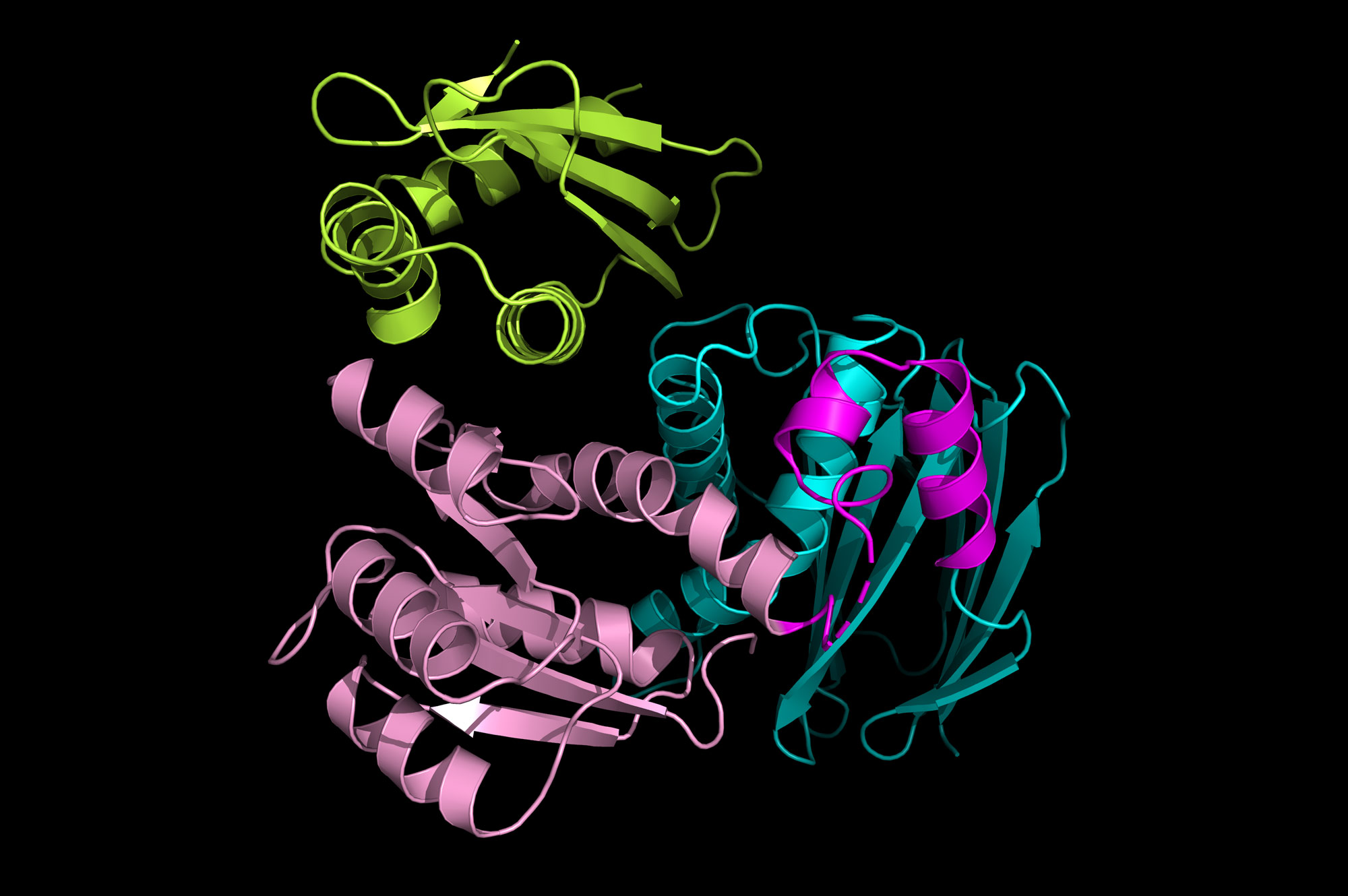
Deaconescu’s team used X-ray crystallography to determine the structure of IraD bound to RssB at atomic-level resolution. The method involves crystallizing a protein or protein complex and observing how the crystals scatter x-rays. The RssB–IraD complex looks somewhat like a trefoil where RssB forms two lobes that both contact IraD in a “hug.” Deaconescu thinks it is likely IraD interacts with one lobe, or domain, of RssB first and then the other domain closes in, helping to form a stable complex.
“We initially thought that IraD might only bind with one domain, but instead it binds to both domains and locks them together in a closed conformation,” Deaconescu said. “That was the biggest surprise about the structure.”
Deaconescu’s collaborators at the National Cancer Institute studied the interaction of RssB and IraD genetically and in laboratory tests. They selectively mutated key regions involved in the interaction. They confirmed that the regions identified in the structure were important for the interaction of the two proteins in bacterial cells and for their activity in laboratory tests involving the cellular recycling machinery.
In the future, Deaconescu plans to study the other inhibition factors that recognize RssB. She thinks that other inhibition factors, which are triggered by different stress signals such as the depletion of various nutrients, may bind to RssB in different ways. They might bind to different parts of the protein or even recognize different conformations as RssB is a very flexible and dynamic protein, more like a multi-purpose hand than a rigid lock with only one key.
Deaconescu is also interested in what happens when the stress that triggers the inhibition factor is resolved and the bacteria need to return to recycling the master regulator.
In addition to Deaconescu, other authors include former research assistant Victoria Dorich, postdoctoral fellows Christiane Brugger and Margaret Suhanovsky, and undergraduate Amita Sastry from Brown as well as Arti Tripathi, Joel Hoskins, Song Tong, Sue Wickner and Susan Gottesman from the National Cancer Institute.
The National Institutes of Health (R01 GM121975) and a Salomon Research Award from Brown University supported the research.