How is the complexity of the nervous system achieved with a surprisingly small number of genes?
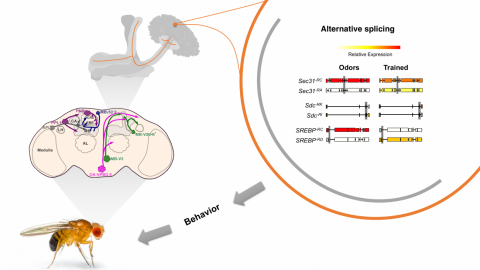
The observation that alternative splice isoforms are enriched in brain tissue from flies to humans has led to the hypothesis that alternative splicing provides the molecular diversity necessary for brain formation and function.
Our cell-specific transcriptomics reveal significant splice isoform diversity between behavioral conditions. And Professors Karla Kaun and Kate O’Connor-Giles hypothesize that this isoform switching induces neuroadaptations in reward circuits that increase vulnerability to addiction.
However, the observation of splicing differences does not address whether the alternative transcript isoform usage has functional consequences. With the exception of a small number of intriguing examples, the functional relevance of alternative splicing in the brain is not known. Kaun and O’Connor-Giles are working on a scalable functional approach to fill this void and gain fundamental biological insights into the dynamic regulation of alternative splicing in the brain.